
The Global Health Drug Discovery Institute (opens in new tab) (GHDDI) and Microsoft Research have reached a milestone in tuberculosis (TB) drug research with TamGen (opens in new tab), an open-source (opens in new tab), transformer-based chemical language model for developing target-specific drug compounds. Working in collaboration, the joint team successfully identified several promising inhibitors for a TB protease, with the most effective compound showing significant bioactivity. Research shows that TamGen can also optimize existing molecules by designing target-aware molecule fragments, potentially enabling the discovery of novel compounds that build on a known molecular core structure.
Generative AI helps overcome limitations in drug discovery
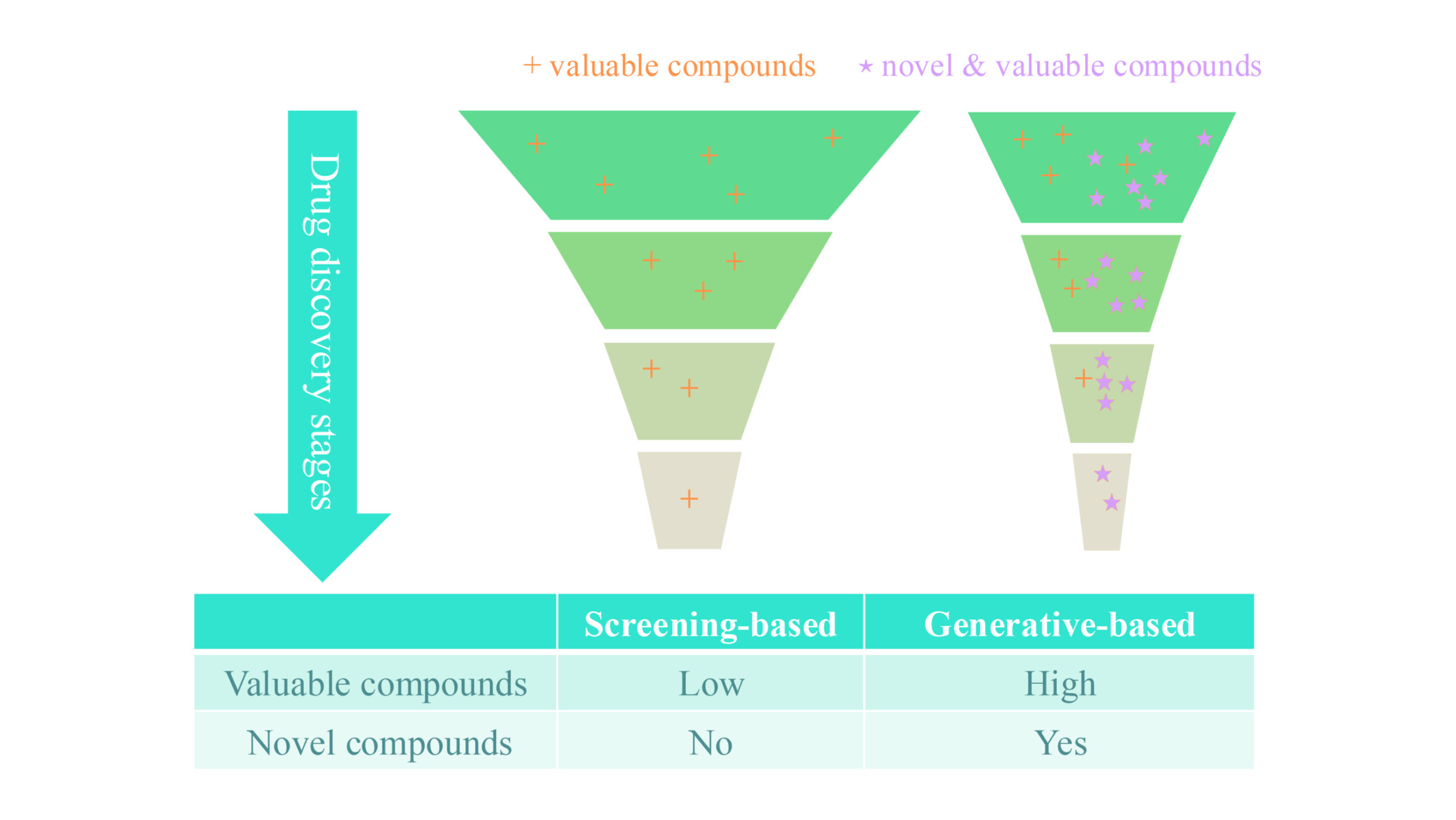
Generative AI is opening new avenues for scientific exploration by allowing computers to autonomously learn and produce original content. TamGen offers a new approach to drug discovery by applying the principles of generative AI to molecular design. Unlike traditional methods, which depend on systematically screening known compounds—a process that is long, complex, and costly due to its reliance on empirical knowledge and the time-consuming task of exploring a vast chemical library—generative AI provides opportunities for designing entirely new chemical structures.
TamGen goes beyond analyzing existing data by generating chemically diverse compounds that conventional approaches might miss. Figure 1 shows that generative AI expands chemical exploration, allowing for a deeper and more comprehensive search for therapeutic solutions compared to traditional methods.
TamGen workflow
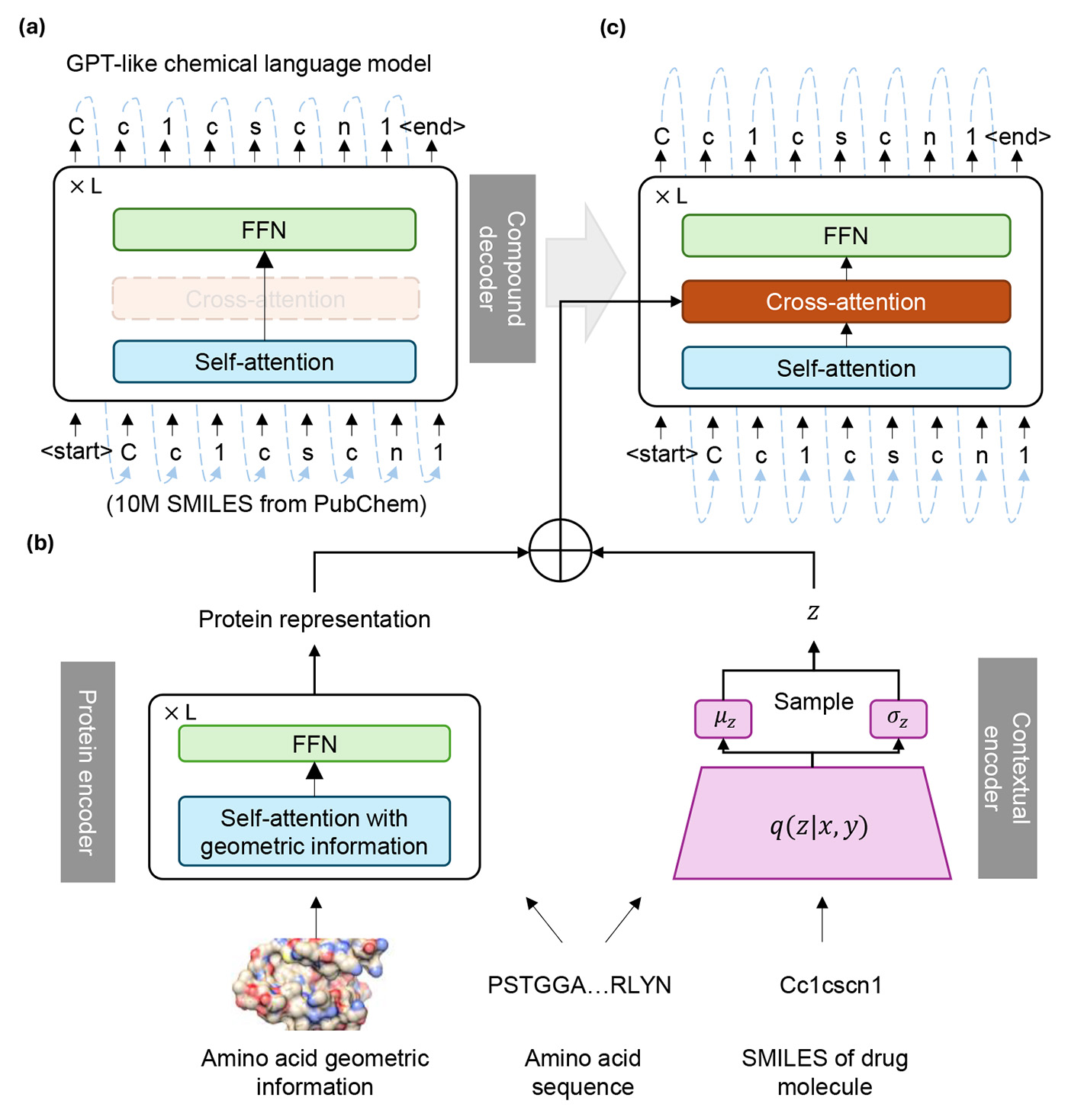
TamGen’s workflow uses generative AI to design target-specific chemical compounds. Building on the success of large language models (LLMs), we adapted a similar approach for molecular generation, using a training method like that of GPT models, which involves next-token prediction. Molecules were first converted into a simplified molecular input line entry system (SMILES)—a notation representing molecular structures as symbol sequences, similar to text. We then developed a protein encoder to process information about proteins, including their 3D structure.
A contextual encoder combines insights from medical professionals with data on the protein target and existing compounds that have proven to be effective or promising. Using expert knowledge and computational analysis, this encoder guides the compound generator to produce new molecules that are more likely to bind to a given protein. This workflow is illustrated in Figure 2.
Evaluating TamGen computationally
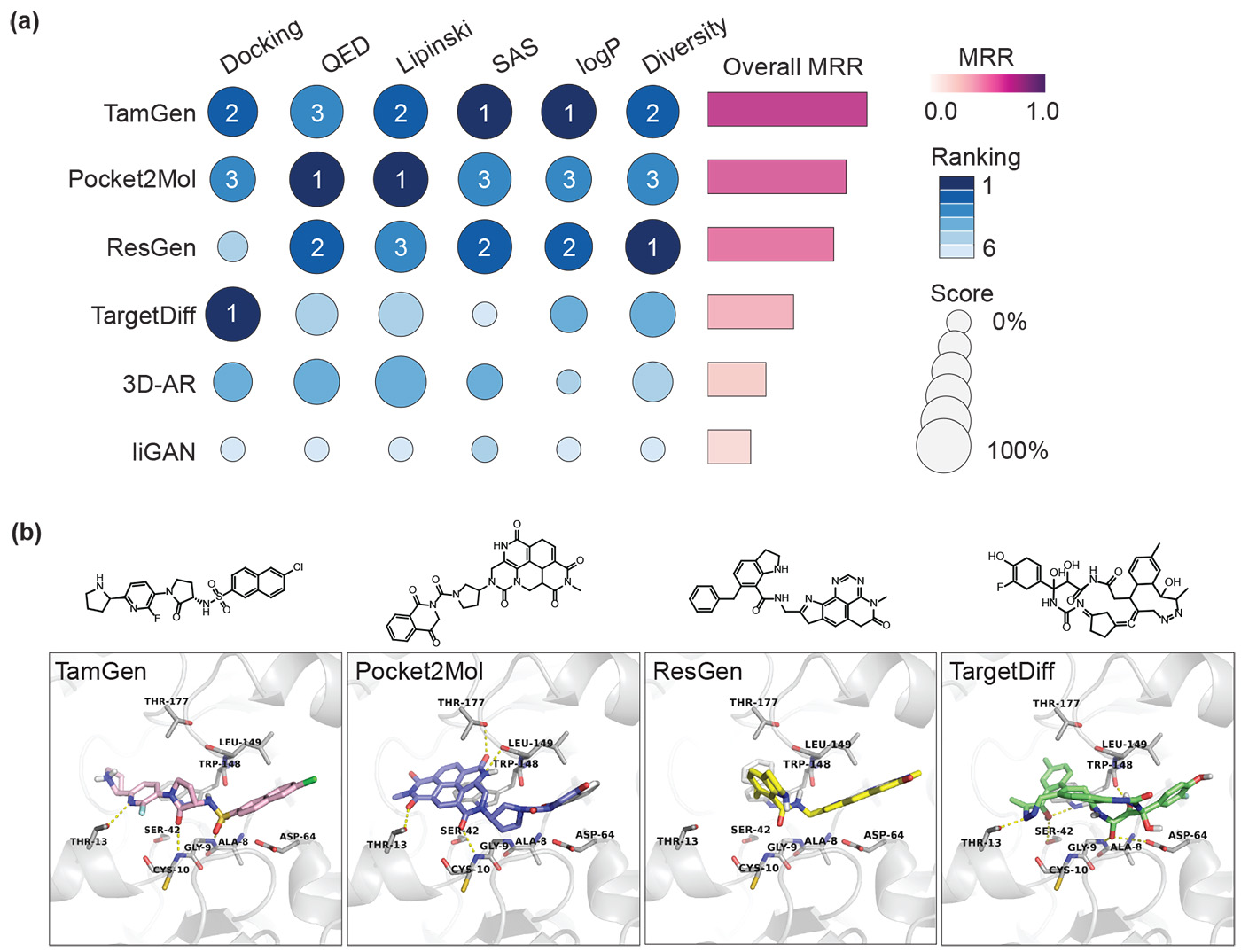
To evaluate TamGen’s performance, we compared it to five other common methods used to create 3D shapes of molecules intended to bind to certain proteins. We evaluated these methods using the CrossDocked benchmark, a dataset used in AI research to assess the quality of molecule generation conditioned on a target protein.
Evaluation metrics:
- Docking score: Measures how well a molecule binds to a target protein.
- Quantitative estimate of drug-likeness (QED): Assesses how good a candidate a molecule is for a drug.
- Synthesis accessibility score (SAS): Measures how easy or difficult it is to synthesize a particular chemical compound in a lab.
- Ro5 (Lipinski’s rule of five): Determines how likely a compound can be developed into an oral drug.
- LogP: Tests a compound’s ability to move between water and fats.
- Diversity: Measures the range of different molecular structures and properties in a collection of compounds.
The findings, illustrated in Figure 3, show TamGen’s overall performance. While other methods may produce compounds that bind more strongly, they often include multiple interconnected ring structures. Research indicates that more of these structures can lower synthesis accessibility (SAS) and increase cellular toxicity, making these compounds harder to develop. We believe that molecular pretraining of the model contributed to the overall effectiveness of the compounds TamGen generated.
Experimental lab verification
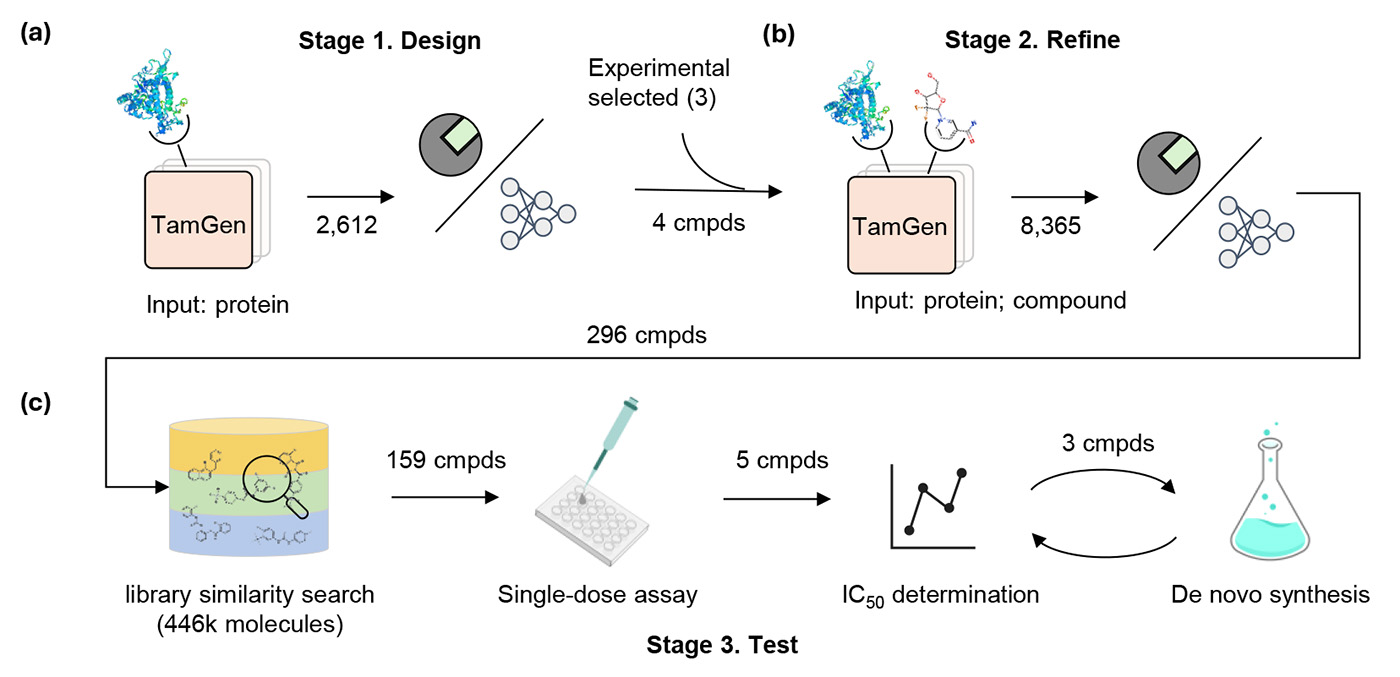
To ensure real-world applicability, we also validated our findings in a hands-on lab environment. Here, we focused on the ClpP protease in Mycobacterium tuberculosis as the target because it plays a significant role in the bacterium’s survival under stress conditions. We proposed the Design-Refine-Test pipeline to effectively identify molecular compounds for TB drug discovery.
Design stage: We began by using TamGen to analyze the binding pocket of the protease, where molecules can attach and influence its function. TamGen generated about 2,600 potential compounds that could fit into this pocket. We assessed these compounds based on how well they could attach to the protease and their predicted biological effects, narrowing it down to four promising candidates.
Refine stage: Next, we entered the four compounds into TamGen, along with three molecular fragments that had been validated in previous lab experiments. This generated a total of 8,600 new compounds, which we screened again using the same criteria, eventually narrowing the selection to 296 compounds.
Test stage: Because synthesizing all 296 compounds wasn’t feasible, we identified similar compounds available in commercial libraries and tested their initial activity against TB. Five compounds showed promising results. We then synthesized one of the originals and two variants of another. Additionally, we categorized the generated compounds into clusters, selected the top 10% from each cluster based on docking scores, and after manual review, synthesized eight more compounds.
The team from Microsoft Research generated the compounds by TamGen, and the GHDDI team conducted binding analysis, structure–activity relationship studies, and lab experiments to verify the compounds’ inhibitory effect on the ClpP protease, measuring their capacity to interfere with or reduce its activity. Lower IC50 values signify greater potency. Out of the 16 compounds tested, 14 showed strong inhibitory activity measuring under 40 µM, indicating high potential. The most effective compound had a measured IC50 value of 1.88 µM.
From molecule to fragment generation
In addition to generating new molecules, TamGen can optimize existing ones by designing smaller molecular fragments. In this fragment generation process, TamGen builds on a given protein target and a molecular core structure to design new compounds around that core. By incorporating information about the target protein, it generates fragments that are highly specific to the target. This approach moves beyond traditional methods that rely on pre-existing databases, which often limit both novelty and effectiveness of molecular fragments.
For fragment generation, we adjusted the input to TamGen’s compound generator. We modified the SMILES string to ensure it ended at the desired growth site. This was done by specifying the fragment we wanted to retain and its connection point for further growth. The tailored SMILES string was then fed into the compound generator to extend the molecule.
We evaluated this method by targeting the ClpP protease for TB, achieving a more than tenfold improvement in the binding affinity of the generated compound compared to the original. Some compounds also demonstrated slow binding, indicating potential for prolonged action and improved selectivity for the target protein.
AI’s potential in drug discovery
TamGen showcases the transformative potential of generative AI in drug design, combining advanced molecular modeling with researcher-AI collaboration. Tasks that once took years can now be accomplished in a fraction of the time. This research underscores AI’s expanding role in drug discovery and its promise for developing effective treatments against persistent infectious diseases like TB.
Looking ahead, we plan to integrate advanced techniques into TamGen, including diffusion models for generating 3D structures, reinforcement learning to apply physical constraints, and molecular dynamics simulations to capture proteins’ shifting shapes. These enhancements aim to improve how well generated compounds bind to target proteins, increase their ability to be synthesized, and strengthen other critical drug properties.




