Making the Most of Text Semantics to Improve Biomedical Vision-Language Processing
- Benedikt Boecking ,
- Naoto Usuyama ,
- Shruthi Bannur ,
- Daniel Coelho de Castro ,
- Anton Schwaighofer ,
- Stephanie Hyland ,
- Maria Teodora Wetscherek ,
- Tristan Naumann ,
- Aditya Nori ,
- Javier Alvarez-Valle ,
- Hoifung Poon ,
- Ozan Oktay
ABSTRACT: Multi-modal data abounds in biomedicine, such as radiology images and reports. Interpreting this data at scale is essential for improving clinical care and accelerating clinical research. Biomedical text with its complex semantics poses additional challenges in vision-language modelling compared to the general domain, and previous work has used insufficiently adapted models that lack domain-specific language understanding. In this paper, we show that principled textual semantic modelling can substantially improve contrastive learning in self-supervised vision-language processing. We release a language model that achieves state-of-the-art results in radiology natural language inference through its improved vocabulary and novel language pretraining objective leveraging semantics and discourse characteristics in radiology reports. Further, we propose a self-supervised joint vision–language approach with a focus on better text modelling. It establishes new state of the art results on a wide range of publicly available benchmarks, in part by leveraging our new domain-specific language model. We release a new dataset with locally aligned phrase grounding annotations by radiologists to facilitate the study of complex semantic modelling in biomedical vision-language processing. A broad evaluation, including on this new dataset, shows that our contrastive learning approach, aided by textual-semantic modelling, outperforms prior methods in segmentation tasks, despite only using a global-alignment objective.
Motivation
Clinical motivation: Growing backlogs of medical image reporting puts pressure on radiologists and leads to errors and omissions.
Scalability: ML models require a vast number of manual annotations (clinicians’ time is precious). Existing models are often limited to a fixed set of abnormalities or body-part.
Domain-specific challenges: Lack of foundation models suitable for health data (e.g., image and text), smaller scale datasets, domain specific-language.
Approach
CXR-BERT language model
CXR-BERT-General is a language encoder model trained specifically with biomedical text data (e.g., PubMed abstracts, and MIMIC clinical notes) to learn domain specific vocabulary and semantics. In the proposed framework, this canonical model is continually pre-trained on MIMIC-CXR dataset to specialise to chest X-ray radiology reports via masked language modelling (MLM), contrastive learning and text augmentations (sentence shuffle). We have made two models available on Hugging Face at https://aka.ms/biovil-models (opens in new tab):
- The new chest X-ray (CXR) domain-specific language model, CXR-BERT-specialized (opens in new tab) (Fig. 1)
- A canonical model that can be used for other applications, CXR-BERT-general (opens in new tab)
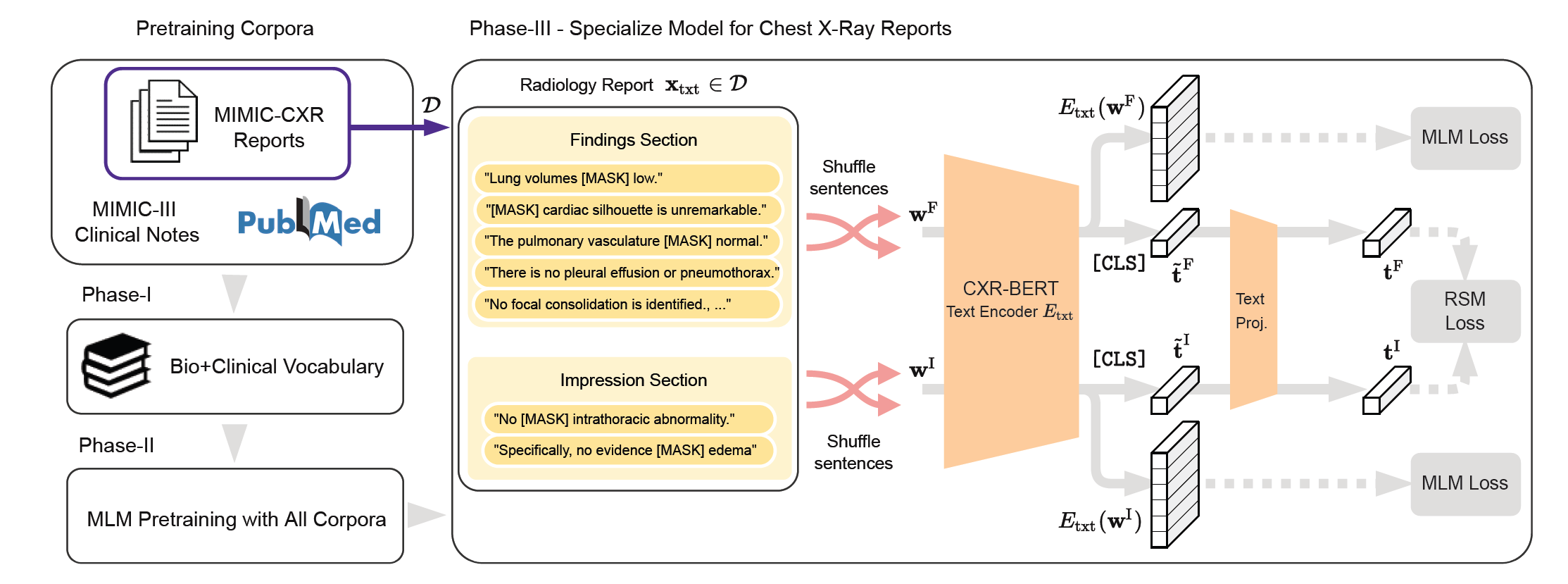
Figure 1: The proposed CXR-BERT text encoder has three phases of pretraining: (I) Pre-training with biomedical corpora (e.g., PubMed Abstracts, MIMIC-III clinical notes), (II) building a biomedical/clinical vocabulary, and (III) further specialising to chest radiology domain by performing contrastive learning between radiology reports and leveraging text augmentations.
BioViL
A self-supervised Vision-Language Processing (VLP) approach for paired biomedical data (BioViL, Fig.2). https://aka.ms/biovil-code (opens in new tab)
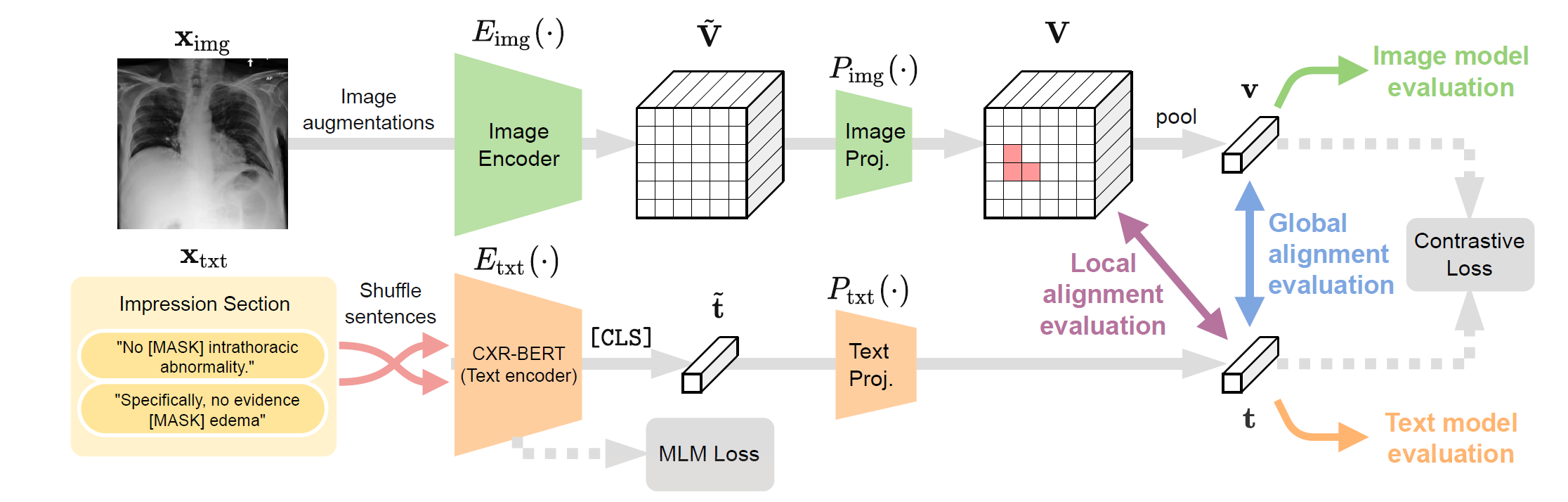
Figure 2: BioViL leverages our radiology-specific text encoder (CXR-BERT) in a multi-modal contrastive learning framework to train image and text encoders that can be aligned in the joint latent space. The proposed learning framework can be coupled with local-contrastive objectives as well.
MS-CXR dataset
MS-CXR is a phrase grounding dataset for chest X-ray data. It allows fine-grained evaluation of joint text-image understanding in a biomedical domain. This dataset was manually annotated and curated by two expert radiologist and comprises 1162 image bounding-box & sentence pairs with 8 different high-level clinical findings.
This dataset is released on PhysioNet: https://aka.ms/ms-cxr (opens in new tab)
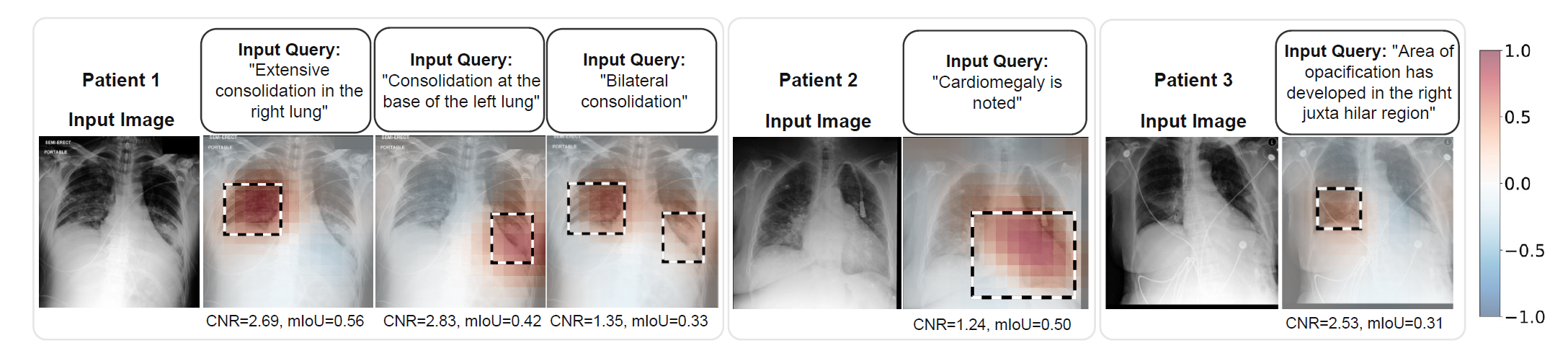
Figure 3: Examples from the MS-CXR dataset. The overlaid colormap is showing cosine similarities of the embeddings obtained from image patches and radiology sentences, where the red colour represents better agreement between the two modalities.
Getting started
The best way to get started is by running the phrase grounding notebook (opens in new tab). All the dependencies will be installed upon execution, so Python 3.7 and Jupyter (opens in new tab) are the only requirements to get started.
The notebook can also be run on Binder (opens in new tab), without the need to download any code or install any libraries:
Resources:
- HI-ML Multimodal Toolkit (opens in new tab), a Python library for multi-modal learning for healthcare and life sciences.
- The multimodal toolkit is part of the more general HI-ML Open-source Toolkit that helps to simplify deep learning models for healthcare and life sciences.
- We have made two models available on Hugging Face at https://aka.ms/biovil-models (opens in new tab)
- The new chest X-ray (CXR) domain-specific language model, CXR-BERT-specialized (opens in new tab) (Fig. 1)
- A canonical model that can be used for other applications, CXR-BERT-general (opens in new tab)
- MS-CXR dataset is released on PhysioNet: https://aka.ms/ms-cxr (opens in new tab)
Acknowledgements
We would like to thank Dr Javier González and Fernando Pérez-Garcia for their valuable feedback and contributions, Hannah Richardson for helping with the compliance review of the datasets, Dr Matthew Lungren for their clinical input and data annotations provided to this study, and lastly Dr Kenji Takeda for preparing the web-content, helping with its presentation and supporting the HI-ML OSS program.
 GitHub
GitHub
Related Tools
Biomedical Visual-Language Processing (BioViL)
February 7, 2023
BioViL is a machine learning model trained on biomedical vision and language datasets at scale. It does not require manual annotations and can leverage historical raw clinical image acquisitions and clinical notes.
Temporal Vision-Language Processing (BioViL-T)
March 24, 2023
BioViL-T is a Vision-Language model trained on sequences of biomedical image and text data at a scale. It does not require manual annotations and can leverage historical raw clinical image acquisitions and clinical notes. The temporal extension enables image and text encoders to become sensitive to disease progression information present in existing datasets. See Learning to Exploit Temporal Structure for Biomedical Vision-Language Processing - Microsoft Research for further information.
Making the Most of Text Semantics to Improve Biomedical Vision-Language Processing
Presented by the Microsoft Health Futures team at ECCV 2022 – Poster session #5
