The use of self-supervision from image-text pairs has been a key enabler in the development of scalable and flexible vision-language AI models in not only general domains but also in biomedical domains such as radiology. The goal in the radiology setting is to produce rich training signals without requiring manual labels so the models can learn to accurately recognize and locate findings in the images and relate them to content in radiology reports.
Radiologists use radiology reports to describe imaging findings and offer a clinical diagnosis or a range of possible diagnoses, all of which can be influenced by considering the findings on previous imaging studies. In fact, comparisons with previous images are crucial for radiologists to make informed decisions. These comparisons can provide valuable context for determining whether a condition is a new concern or improving, deteriorating, or stable if an existing condition and can inform more appropriate treatment recommendations. Despite the importance of comparisons, current AI solutions for radiology often fall short in aligning images with report data because of the lack of access to prior scans. Current AI solutions also typically fail to account for the chronological progression of disease or imaging findings often present in biomedical datasets. This can lead to ambiguity in the model training process and can be risky in downstream applications such as automated report generation, where models may make up temporal content without access to past medical scans. In short, this limits the real-world applicability of such AI models to empower caregivers and augment existing workflows.
In our previous work, we demonstrated that multimodal self-supervised learning of radiology images and reports can yield significant performance improvement in downstream applications of machine learning models, such as detecting the presence of medical conditions and localizing these findings within the images. In our latest study, which is being presented at the 2023 IEEE/CVF Computer Vision and Pattern Recognition Conference (CVPR), we propose BioViL-T, a self-supervised training framework that further increases the data efficiency of this learning paradigm by leveraging the temporal structure present in biomedical datasets. This approach enables the incorporation of temporal information and has the potential to perform complementary self-supervision without the need for additional data, resulting in improved predictive performance.
Our proposed approach can handle missing or spatially misaligned images and can potentially scale to process a large number of prior images. By leveraging the existing temporal structure available in datasets, BioViL-T achieves state-of-the-art results on several downstream benchmarks. We’ve made both our models (opens in new tab) and source code (opens in new tab) open source, allowing for a comprehensive exploration and validation of the results discussed in our study. We’ve also released a new multimodal temporal benchmark dataset, MS-CXR-T (opens in new tab), to support further research into longitudinal modeling of medical images and text data.
Microsoft research podcast
Connecting the data points
Solving for the static case in vision-language processing—that is, learning with pairs of single images and captions—is a natural first step in advancing the field. So it’s not surprising that current biomedical vision-language processing work has largely focused on tasks that are dependent on features or abnormalities present at a single point in time—what is a patient’s current condition, and what is a likely diagnosis?—treating image-text pairs such as x-rays and corresponding reports in today’s datasets as independent data points. When prior imaging findings are referenced in reports, that information is often ignored or removed in the training process. Further, a lack of publicly available datasets containing longitudinal series of imaging examinations and reports has further challenged the incorporation of temporal information into medical imaging benchmarks.
Thanks to our early and close collaboration with practicing radiologists and our long-standing work with Nuance, a leading provider of AI solutions in the radiology space that was acquired by Microsoft in 2022 (opens in new tab), we’ve been able to better understand clinician workflow in the radiological imaging setting. That includes how radiology data is created, what its different components are, and how routinely radiologists refer to prior studies in the context of interpreting medical images. With these insights, we were able to identify temporal alignment of text across multiple images as a clinically significant research problem. To ground, or associate, report information such as “pleural effusion has improved compared to previous study” with the imaging modality requires access to the prior imaging study. We were able to tackle this challenge without gathering additional data or annotations.
As an innovative solution, we leveraged the metadata from de-identified public datasets like MIMIC-CXR (opens in new tab). This metadata preserves the original order and intervals of studies, allowing us to connect various images over time and observe disease progression. Developing more data efficient and smart solutions in the healthcare space, where data sources are scarce, is important if we want to develop meaningful AI solutions.
Addressing the challenges of longitudinal analysis
With current and prior images now available for comparison, the question became, how can a model reason about images coming from different time points? Radiological imaging, especially with planar techniques like radiographs, may show noticeable variation. This can be influenced by factors such as the patient’s posture during capture and the positioning of the device. Notably, these variations become more pronounced when images are taken with longer time gaps in between. To manage variations, current approaches to longitudinal analysis, largely used for fully supervised learning of image models only, require extensive preprocessing, such as image registration, a technique that attempts to align multiple images taken at different times from different viewpoints. In addition to better managing image variation, we wanted a framework that could be applied to cases in which prior images weren’t relevant or available and the task involved only one image.
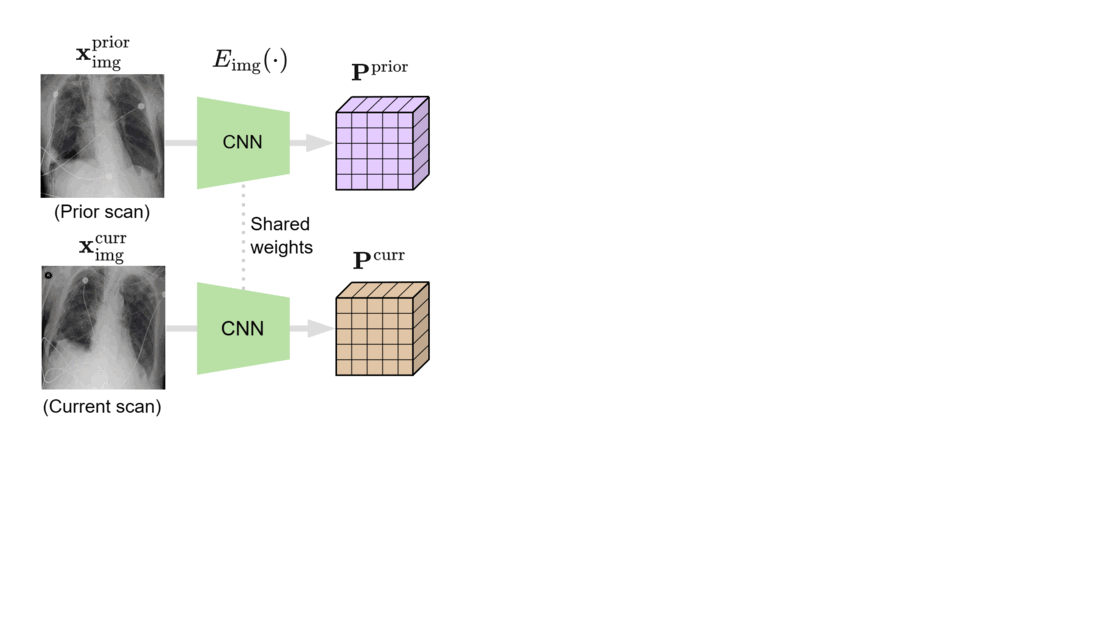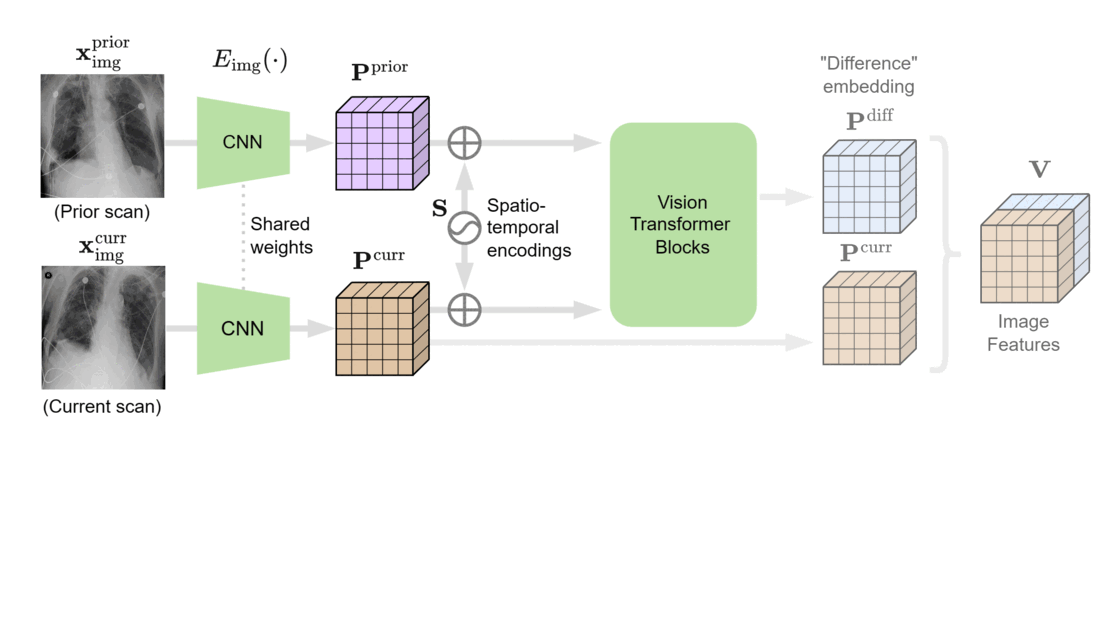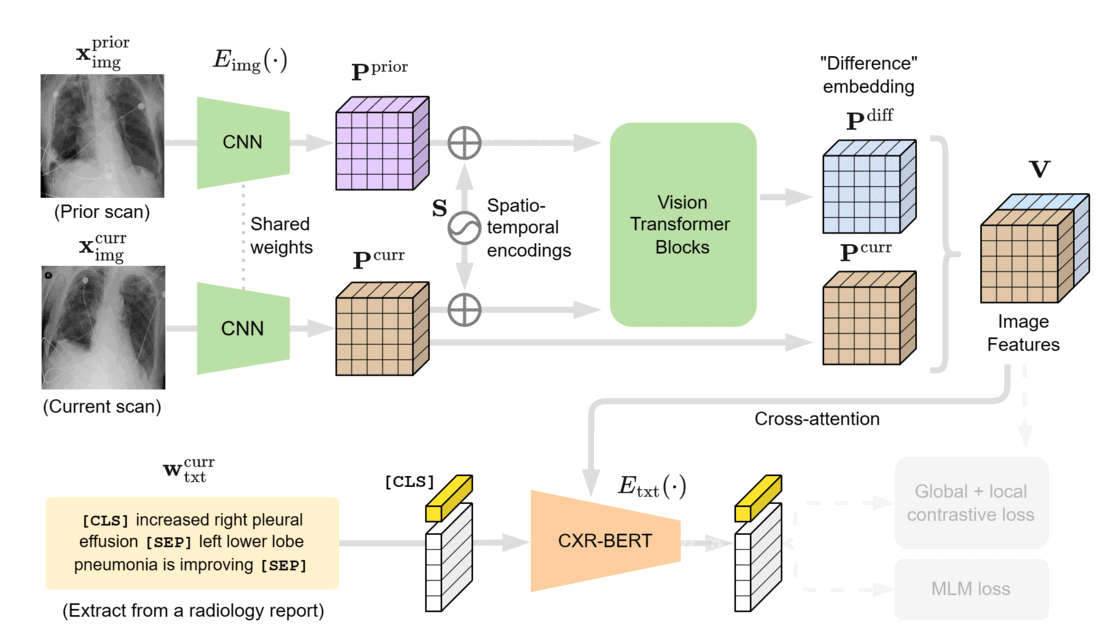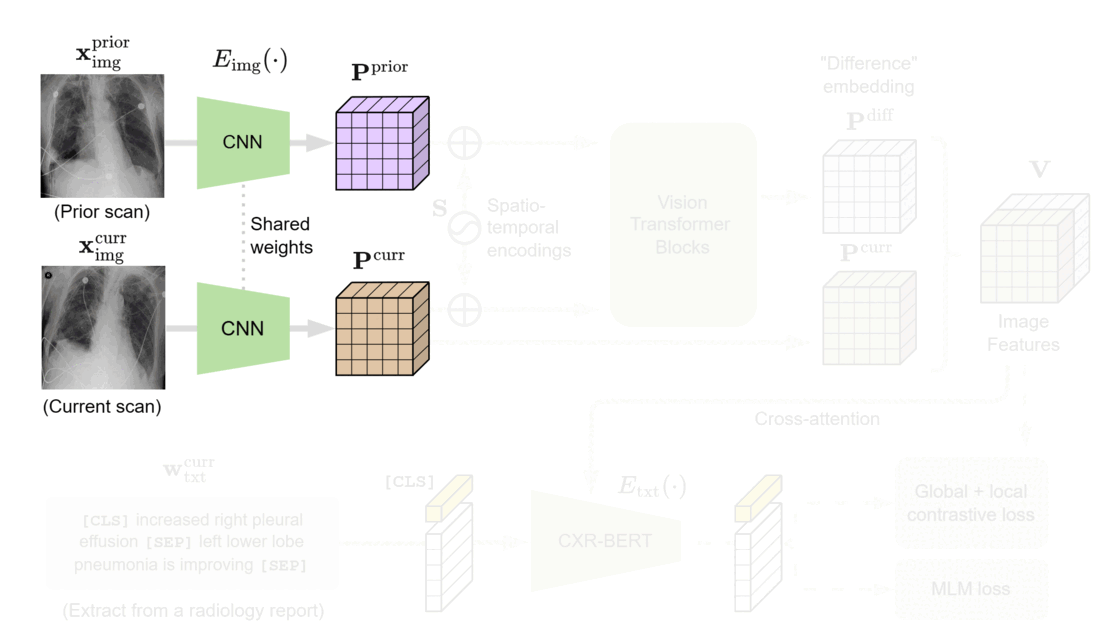
We designed BioViL-T with these challenges in mind. Its main components are a multi-image encoder, consisting of both a vision transformer and a convolutional neural network (CNN), and a text encoder. As illustrated in Figure 1, in the multi-image encoder, each input image is first encoded with the CNN model to independently extract findings, such as opacities, present in each medical scan. Here, the CNN counteracts the large data demands of transformer-based architectures through its efficiency in extracting lower-level semantic features.
At the next stage, the features across time points are matched and compared in the vision transformer block, then aggregated into a single joint representation incorporating both current and historical radiological information. It’s important to note that the transformer architecture can adapt to either single- or multi-image scenarios, thereby better handling situations in which past images are unavailable, such as when there’s no relevant image history. Additionally, a cross-attention mechanism across image regions reduces the need for extensive preprocessing, addressing potential variations across images.
In the final stage, the multi-image encoder is jointly trained with the text encoder to match the image representations with their text counterparts using masked modeling and contrastive supervision techniques. To improve text representations and model supervision, we utilize the domain-specific text encoder CXR-BERT-general (opens in new tab), which is pretrained on clinical text corpora and built on a clinical vocabulary.
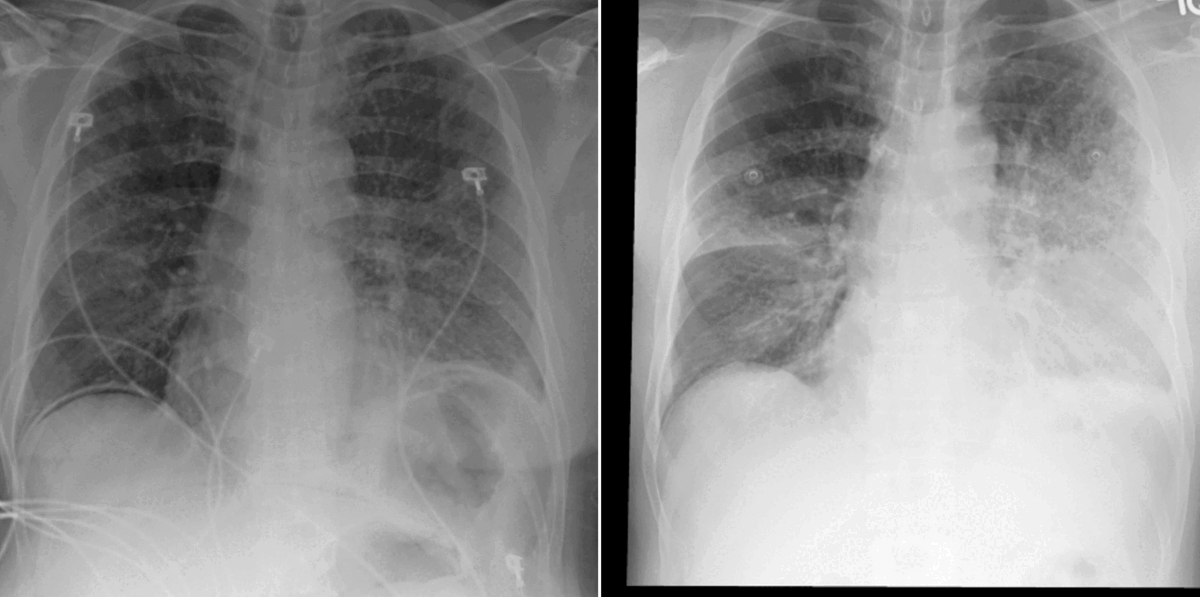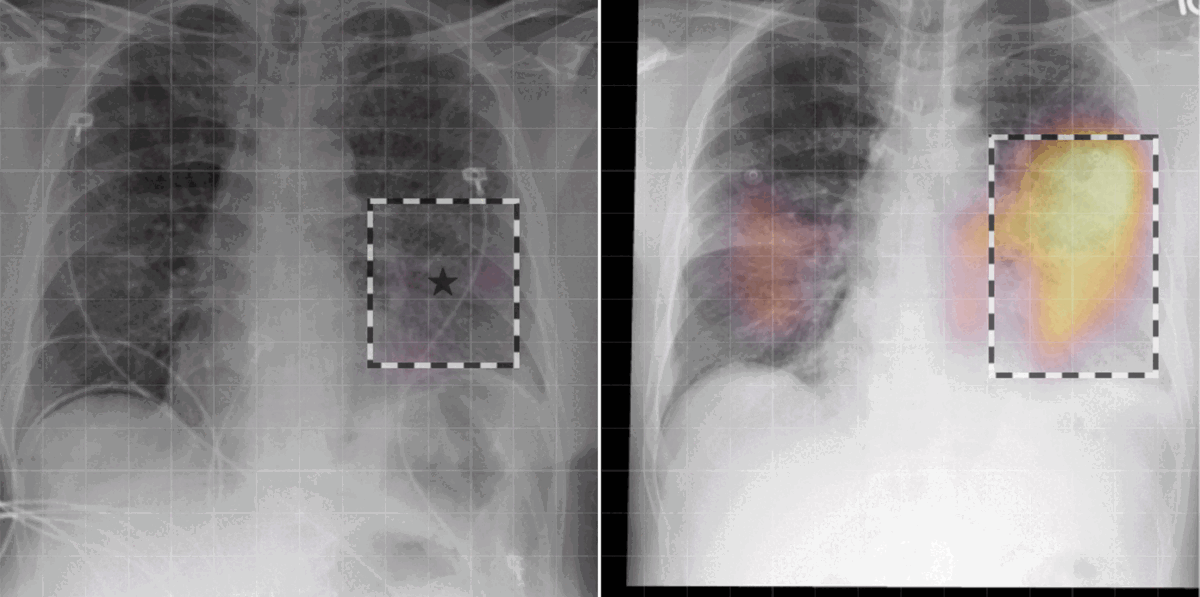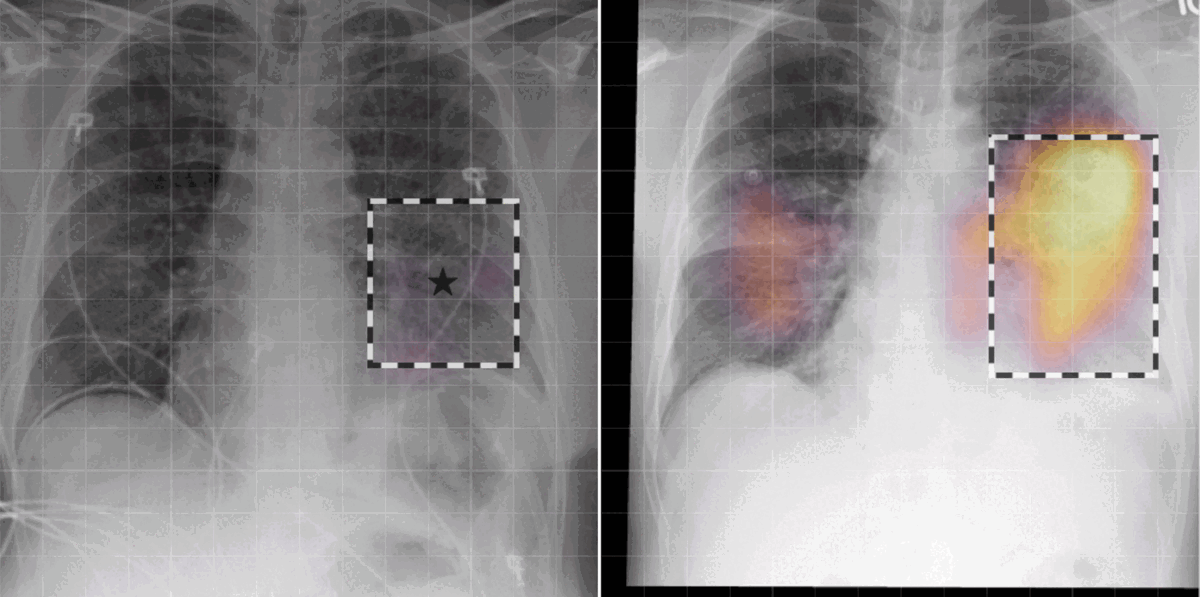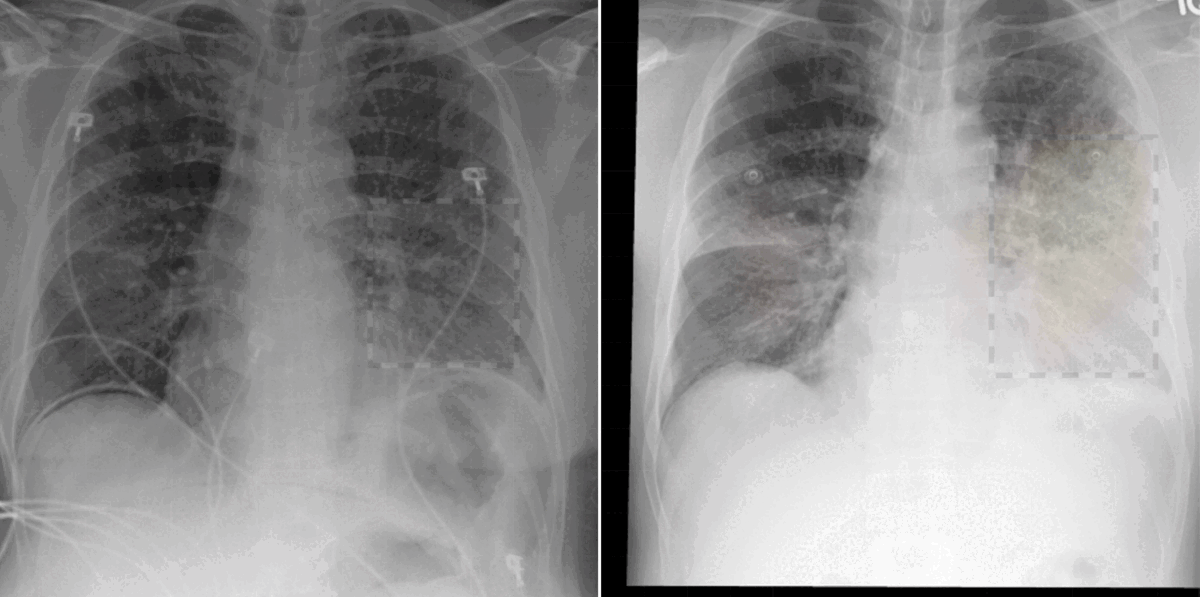
Grounded model prediction
In our work, we found that linking multiple images during pretraining makes for both better language and vision representations, enabling the AI model to better associate information present in both the text and the images. This means that when given a radiology report of a chest x-ray, for example, with the description “increased opacities in the left lower lung compared with prior examination,” a model can more accurately identify, locate, and compare findings, such as opacities. This improved alignment between data modalities is crucial because it allows the model to provide more accurate and relevant insights, such as identifying abnormalities in medical images, generating more accurate diagnostic reports, or tracking the progression of a disease over time.
Two findings were particularly insightful for us during our experimentation with BioViL-T:
- Today’s language-generating AI models are often trained by masking portions of text and then prompting them to fill in the blanks as a means of encouraging the models to account for context in outputting a prediction. We extended the traditional masked language modeling (MLM) approach to be guided by multi-image context, essentially making the approach multimodal. This, in return, helped us better analyze whether BioViL-T was learning a progression based on provided images or making a random prediction of the masked words based solely on the text context. We gave the model radiology images and reports with progression-related language, such as “improving,” masked. An example input would be “pleural effusion has been [MASKED] since yesterday.” We then tasked the model with predicting the missing word(s) based on single and multi-image inputs. When provided with a single image, the model was unsuccessful in completing the task; however, when provided with a current and prior image, performance improved, demonstrating that the model is basing its prediction on the prior image.
- Additionally, we found that training on prior images decreases instances of the generative AI model producing ungrounded outputs that seem plausible but are factually incorrect, in this case, when there’s a lack of information. Prior work into radiology report generation utilizes single input images, resulting in the model potentially outputting text that describes progression without having access to past scans. This severely limits the potential adoption of AI solutions in a high-stakes domain such as healthcare. A decrease in ungrounded outputs, however, could enable automated report generation or assistive writing in the future, which could potentially help reduce administrative duties and ease burnout in the healthcare community. Note that these models aren’t intended for any clinical use at the moment, but they’re important proof points to assess the capabilities of healthcare AI.
Moving longitudinal analysis forward
Through our relationships with practicing radiologists and Nuance, we were able to identify and concentrate on a clinically important research problem, finding that accounting for patient history matters if we want to develop AI solutions with value. To help the research community advance longitudinal analysis, we’ve released a new benchmark dataset. MS-CXR-T (opens in new tab), which was curated by a board-certified radiologist, consists of current-prior image pairs of chest x-rays labeled with a state of progression for the temporal image classification task and pairs of sentences about disease progression that are either contradictory or capture the same assessment but are phrased differently for the sentence similarity task.
We focused on chest x-rays and lung diseases, but we see our work as having the potential to be extended into other medical imaging settings where analyzing images over time plays an important part in clinician decision-making, such as scenarios involving MRI or CT scans. However far the reach, it’s vital to ensure that models such as BioViL-T generalize well across different population groups and under the various conditions in which medical images are captured. This important part of the journey requires extensive benchmarking of models on unseen datasets. These datasets should widely vary in terms of acquisition settings, patient demographics, and disease prevalence. Another aspect of this work we look forward to exploring and monitoring is the potential role of general foundation models like GPT-4 in domain-specific foundation model training and the benefits of pairing larger foundation models with smaller specialized models such as BioViL-T.
To learn more and to access our text and image models and source code, visit the BioViL-T Hugging Face page (opens in new tab) and GitHub (opens in new tab).
Acknowledgments
We’d like to thank our co-authors: Shruthi Bannur, Stephanie Hyland, Qianchu Liu, Fernando Pérez-García, Maximilian Ilse, Daniel C. Castro, Benedikt Boecking, Harshita Sharma, Kenza Bouzid, Anja Thieme, Anton Schwaighofer, Maria Wetscherek, and Aditya Nori. We’d also like to thank Hoifung Poon, Melanie Bernhardt, Melissa Bristow, and Naoto Usuyama for their valuable technical feedback and Hannah Richardson for assisting with compliance reviews.
MEDICAL DEVICE DISCLAIMER
BioViL-T was developed for research purposes and is not designed, intended, or made available as a medical device and should not be used to replace or as a substitute for professional medical advice, diagnosis, treatment, or judgment.





